Software
I develop and maintain a variety of bioinformatics software.
smfishHmrf (spatial transcriptomics)
2018
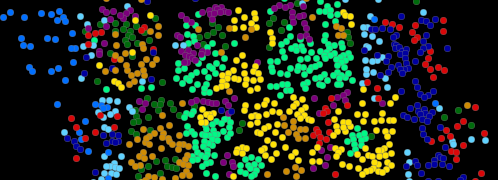
With the first generation seqFISH, we applied it on visual cortex specimen (125 genes).
Accompanying this data, we developed Hidden Markov Random Field (HMRF) to infer spatial domains from this data (bitbucket.org/qzhu/smfish-hmrf/).
Input of HMRF requires only cell coordinate matrix and cell-by-gene expression matrix.
We also have a Support Vector Machine (SVM) based learner to map cell types from a single cell RNAseq dataset onto seqFISH (of same tissue region).
Giotto (spatial transcriptomics)
2020
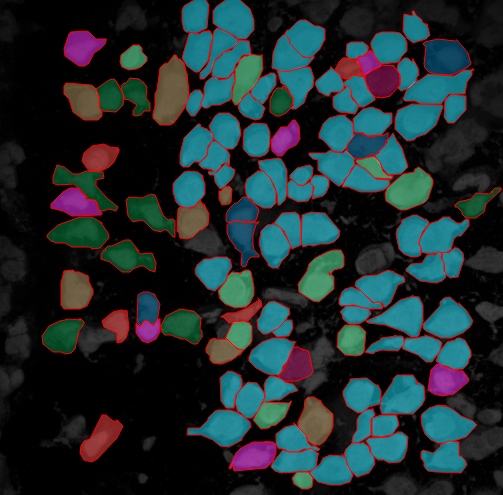
As spatial transcriptomics data get rapidly generated from recent imaging and sequencing-based technologies, a standardized analysis toolkit becomes much in need.
Giotto (spatialgiotto.com) is a set of tools (written in R, Python, Javascript) offering a variety of downstream spatial analyses and interactive visualizations, including spatial clustering, spatial gene detection, cell cell proximity analysis, and histology analysis.
It has been applied to Slide-seq, MERFISH, SeqFISH+, Spatial Transcriptomics, MIBI-TOF, OsmFISH, STARmap.
CUT&RUNTools (epigenomics)
2019
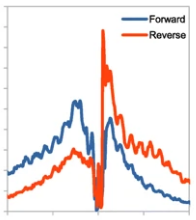
CUT&RUN is a chromatin profiling technology which uses protein-A MNase fusion protein, coupled with an antibody, to cleave around antibody-bound locations on the DNA.
It can be used to map transcription factor and histone binding sites.
This tool (bitbucket.org/qzhudfci/cutruntools) fascilitates data processing, and provides the reads trimming, cut tabulation, motif footprint steps important for the analysis, validation of CUT&RUN.
SEEK (Search based exploration of expression compendia) (big data)
2015
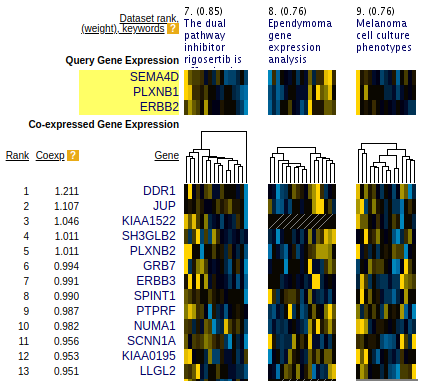
SEEK is a web-server (seek.princeton.edu) that performs coexpression data integration in real-time.
Users enter a query gene or gene-set. SEEK will weight thousands of gene expression datasets (5,000 datasets and 150,000 conditions) based on the query, and integrate these datasets to find robust coexpressed genes.
It visualizes the expression patterns of these genes side-by-side in datasets prioritized to the query genes.